This is the homepage of the research project Multi-scale Modeling and Simulation of Field-Effect Nano-Biosensors. The project proposal was submitted in September 2007 to the call in computational science by the jubilee-fund of the City of Vienna for the Austrian Academy of Sciences (ÖAW). Every second year projects in the natural sciences are funded, and two proposals (out of about 30) were selected for funding in the 2007 computational-science call.
The principal investigator is Clemens Heitzinger with Norbert J. Mauser as co-investigator. The project started in April 2008 at the Wolfgang Pauli Institute (WPI) at the Department of Mathematics at the University of Vienna.
The goal of this project is, simply put, to develop a quantitative theory for modeling and simulation of field-effect nano-biosensors.
Biosensors help us to detect diseases and to detect pathogens. The more we know about which and how many biomolecules there are in our bodies, the better we can respond, even before a disease breaks out. The vision of predictive, personalized, and preemptive medicine means that we try to predict which disease may develop when, that we try to preempt diseases before they occur, and that treatment is available on a personalized basis. To achieve this, we obviously need to know about the biomolecules and pathogens in our bodies, and therefore we need biosensors to detect, e.g., DNA and cancer markers.
Nowadays, biosensors that are used commercially work by labeling the analyte molecules and then actually detecting the labels. This means that the molecules are detected indirectly. Field-effect biosensors, on the contrary, detect the analyte directly by its partial charges (and counter-ions). The idea of field-effect sensors with semiconductor transducers can be traced back to certain pH sensors (ISFETs, Bergveld, 1970s); DNA sensors were demonstrated about 10 years ago (Souteyrand et al., 1997; also Schöning and Poghossian at Jülich); and the subject of field-effect sensors has received attention in nanotechnology recently, when sensors based on nanowires where fabricated (Lieber's group at Harvard, Reed's group at Yale).
Due to the direct detection mechanism, field-effect biosensors have advantages compared to currently employed technology: they are direct, label-free, continuous, and real-time operation. The simpler structure promises cheaper, yet sensitive devices. Portable sensing within a few minutes also makes point-of-care applications possible.

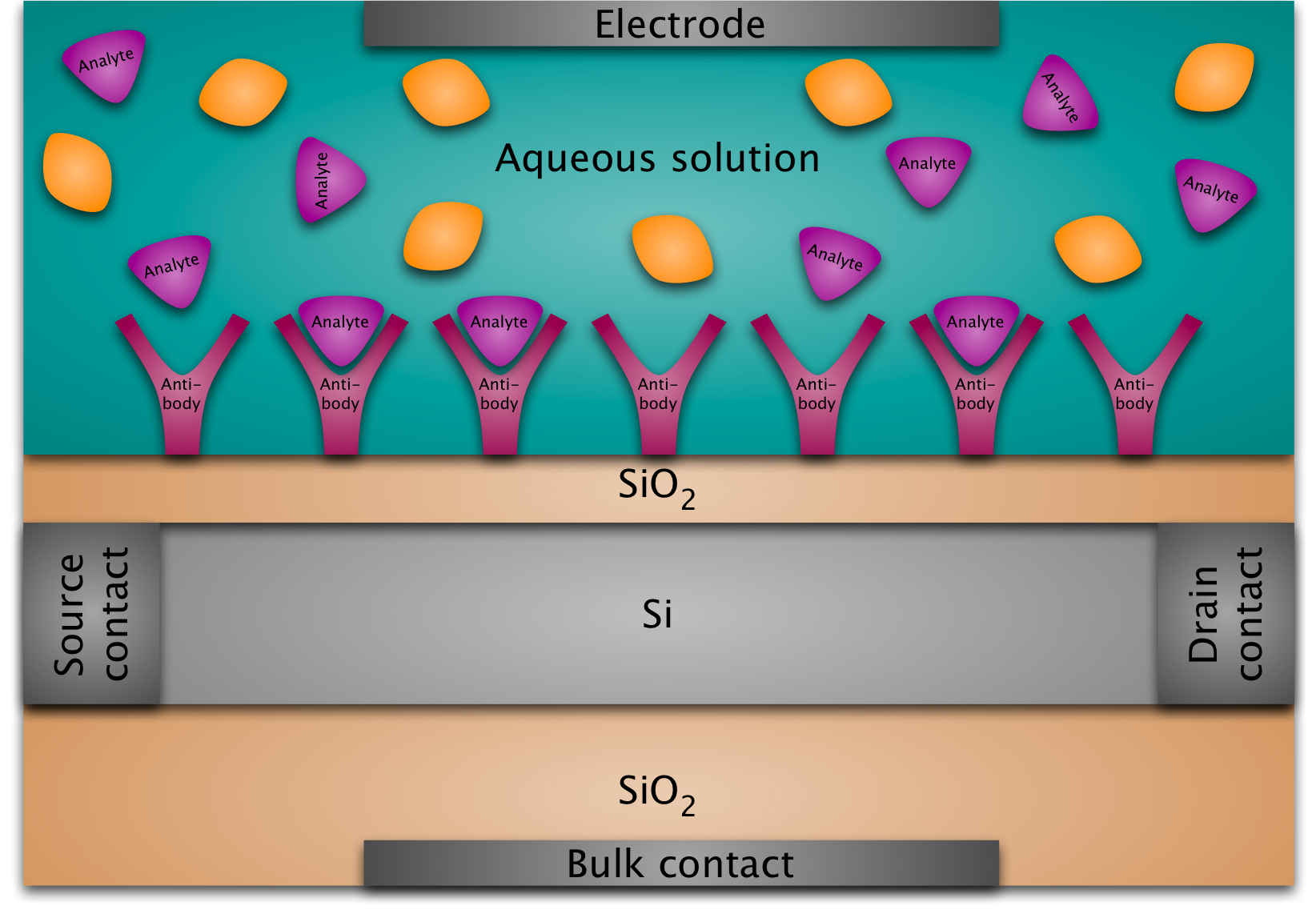
A BioFED (biologically sensitive field-effect device) is basically a MOSFET (metal-oxide-semiconductor field-effect transistor) whose gate structure is replaced by a layer of immobilized molecules at the silicon-oxide surface immersed in an electrolyte. The main idea is that the binding of target molecules to the immobilized probe molecules at the surface modulates the conductance of the semiconductor transducer so that the resulting conductance variation between the two contacts at the left and at the right can be measured and thus provides the detection mechanism.
A DNAFET is a special kind of BioFED: in this case the probe molecules are ssDNA (single-stranded DNA) and the target molecules are the complementary strands. The transducer can be a silicon nanowire on an oxide surface or a silicon nanoplate (possibly a silicon-on-insulator structure).
The aqueous solution in our sensor is an electrolyte, i.e., it contains cations and anions. This means that whenever we put a charge into the solution, the ions in the solution will rearrange to minimize their potential energy. For example, the negative partial charges of the phosphate groups of the DNA backbone will attract positive counter-ions. This is called screening. It implies that the electric potential due to our original charge will be smaller (i.e., it will be screened) after the ions have rearranged. (One could imagine at this point that it would be a good idea to get rid of the ions in the water altogether. This is, however, not feasible in practice. Moreover, a certain amount of ionic concentration is necessary for DNA hybridization.)
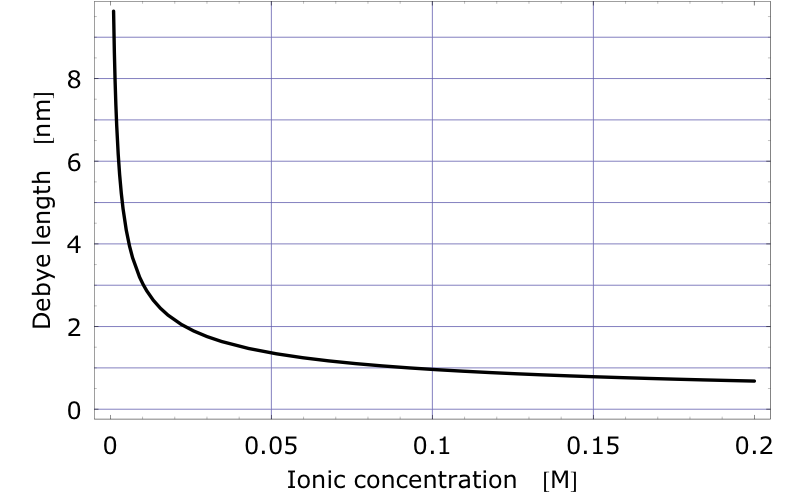
The Debye length can be used to measure the amount of screening. It is the distance at which the electric potential due to a point charge in an electrolyte has dropped off by a certain factor. It depends on the ionic concentration of the liquid and it decreases rapidly as the ionic concentration increases. The figure shows the Debye length of a unit charge as a function of Na+Cl- concentration. Physiologically relevant concentrations correspond to a 150mM or 160mM solution. It can be seen that at the salt concentration of blood or serum the Debye length of a unit charge is less than 1nm.
Screening of course is a fundamental issue for field-effect sensors, since we try to detect partial charges screened by counter-ions. This means that modeling and simulation of field-effect sensors must be caried out carefully including all the charges in the system.
Despite the recent experimental successes, quantitative models (not containing any fitting parameters) for field-effect sensors have been missing. There are probably two reasons: it is a new technology and the modeling is complicated by a few reasons.
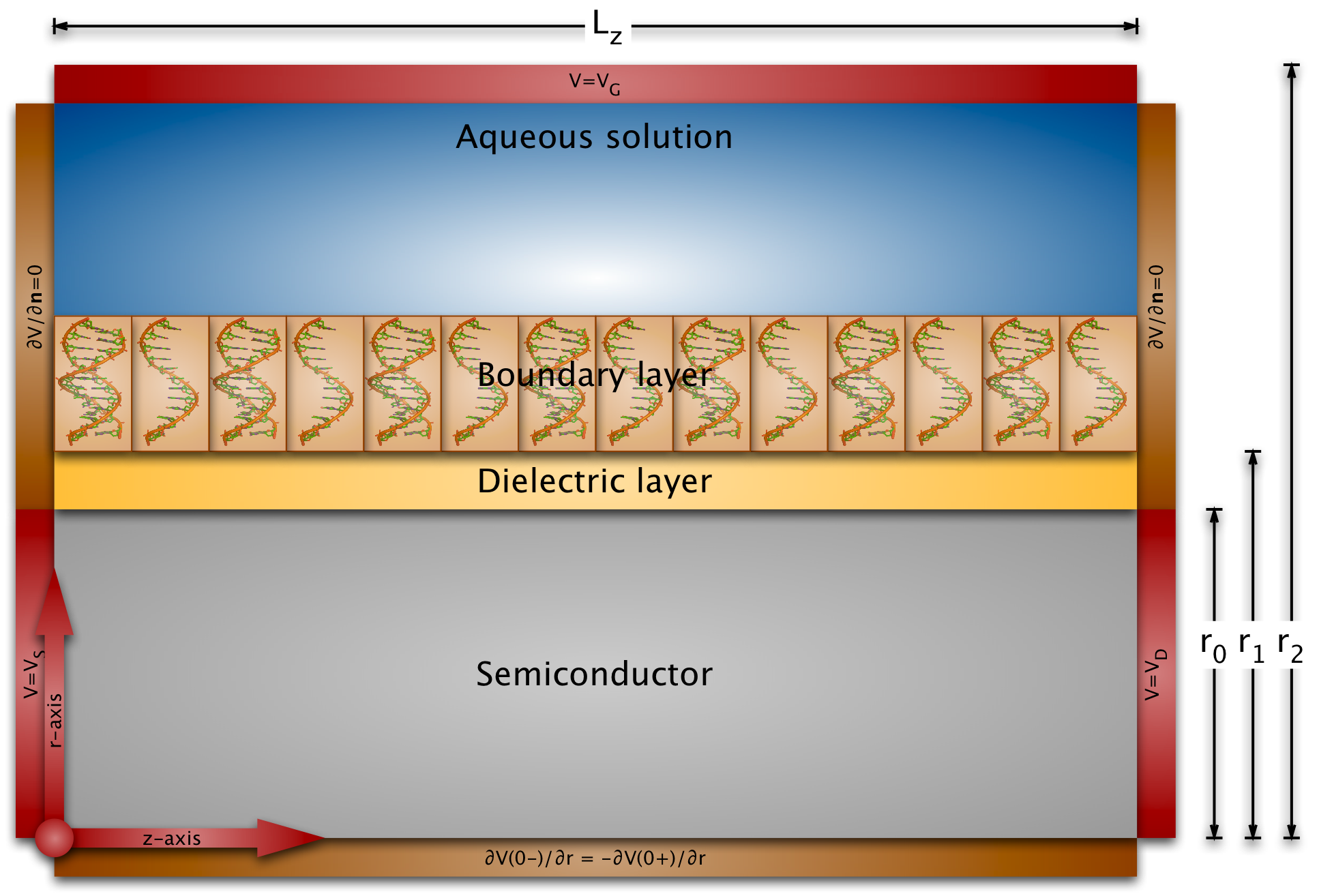
Modeling field-effect sensors must take into account the electrostatics of the liquid, of the probe and target molecules in the boundary layer, the binding efficiency of the probes and targets, and the electrostatics and conductance of the semiconducting transducer.
The length scales of the biomolecules and the dimensions of the sensor surfaces are very different: the partial charge distribution and the electric potential of biomolecules varies on the Angstrom or nanometer length scale (the diameter of the B-DNA helix is 2nm), whereas the exposed sensor areas have dimensions of micrometers (and even centimeter-long nanowires haven been fabricated). This implies that we cannot just use a conventional semiconductor device simulator, because the grid size would be much too large for any practical simulation.
Therefore we have developed multi-scale models for the simulation of nano-plate and nanowire BioFEDs. We have also investigated the crucial boundary layer by Metropolis Monte Carlo simulations in the constant-voltage ensemble and by solving the Poisson-Boltzmann equation. The conductance variations in our simulations are in good quantitative agreement with published experiments. The simulations also show that most of the published experiments can be explained by a conductance change due to a field-effect. (Competing explanations are leakage currents or chemical drift of some part of the sensor over time.)
The Workshop on Nanostructures in Biology and Physics was organized in Vienna from July 21 to July 25, 2008.
A presentation about the project for the general public will be given on October 13, 2008 (details to be announced later).
We are happy to collaborate with Prof. Dezsö Boda (University of Pannonia, Hungary), Prof. Robert Dutton and Dr. Yang Liu (EE and CIS, Stanford), Prof. Robert Eisenberg (Rush University Medical Center), Prof. Christian Ringhofer (Mathematics, ASU), and Prof. Michael Schöning and Dr. Arshak Poghossian (FH Aachen and FZ Jülich).
Publications are available via this homepage.
| Impressum | webmaster |